- Visibility 213 Views
- Downloads 32 Downloads
- Permissions
- DOI 10.18231/j.ijpca.2024.007
-
CrossMark
- Citation
Molecular docking studies for NPACT ligands for the treatment of melanoma skin cancer
Abstract
Background: Approximately 80% of deaths attributable to skin cancer are attributed to melanoma. The main reason for its life-threatening nature is its high tendency to metastasis. The outlook for melanoma patients with distal metastases is grim, as they have a median survival of only six months, despite the utilization of the most advanced treatments now available. The Extracellular signal-regulated kinase (ERK) pathway is the most frequently altered oncogene in melanoma. Phytochemicals are receiving significant recognition due to their minimal toxicity, affordable price, and widespread acceptance as dietary supplements. Preclinical investigations have revealed many cellular and molecular processes via which phytochemicals function in the prevention and treatment of metastatic melanoma.
Objective: In this study, we performed virtual screening of 464 phytoconstituents were obtained from the Naturally Occurring Plant-based Anti-cancer Compound-Activity-Target (NPACT) database to identify novel ERK inhibitors.
Materials and Methods: Virtual screening carried out prediction of drug-likeness and molecular docking studies with 2OJG, protein target related to ERK pathway.
Results: Nine compounds achieved better docking score as compared to co-crystallized ligand. The accuracy of the docking method was checked using re-docking. Picetannol and sulfurentin were identified as potential inhibitors with docking score of -9.2 and -8.2, respectively. These two phytoconstituents also found to be non-carcinogenic which was predicted using a free webtool, Swiss ADME.
Conclusion: Our finding suggests that Piceatannol has promising potential to be further explored as ERK-pathway inhibitor in treatment of melanoma
Introduction
Melanoma is recognized as a highly perilous type of skin tumor, characterized by rapid metastasis, progression, and a significant mortality rate, particularly when diagnosed at an advanced stage. Melanoma and non-melanoma (basal cell carcinoma and squamous cell carcinoma) are the two subtypes of this condition, with melanoma being more dangerous than the latter. Melanocytes are found across several regions of the body, including the skin, eyes, nasal passages, oral and pharyngeal mucosa, as well as the vaginal and anal mucosa.[1]
The Extracellular signal-regulated kinase (ERK) pathway, which is also called the Ras/Raf/Mitogen-activated protein kinase/ERK kinase (MEK)/extracellular-signal-regulated kinase (Ras/Raf/MEK/ERK) signal transduction system, is often turned on in cancer because of a number of abnormal molecular changes that are not under control. This signalling pathway regulates various essential cellular activities such as cell viability, reproduction, movement, and specialization. It is consistently active in lung, colon, pancreatic, kidney, and ovarian malignancies. Moreover, the ERK pathway contains some of the most well-defined oncogenic alterations, such as Ras and B-Raf. ERK plays a crucial role in the signalling pathway that occurs after Ras, Raf, and MEK. It acts as a central hub where several signalling pathways come together to stimulate transcription.[2]
Database of naturally occurring plant-based compounds with anti-cancer properties and their corresponding activities and targets. The Naturally Occurring Plant-based Anti-cancer Compound-Activity-Target (NPACT) database is a meticulously selected collection of natural chemicals produced from plants that demonstrate anti-cancer properties. The dataset has 1574 entries, and each entry has detailed information about the structure, composition (physical, elemental, and topological), type of cancer, cell lines, inhibitory values Half maximal inhibitory concentration, effective concentration, median effective dose (IC50, EC50, ED50), molecular targets, commercial suppliers, and how similar the compounds are to drugs. NPACT focuses exclusively on the study of natural chemicals derived from plants that have anti-cancer properties. NPACT stands out by offering the bioactivities of these natural compounds against various cancer cell lines and their corresponding molecular targets.[3]
The present work aims to investigate the anticancer properties of NPACT ligands against the ERK signalling pathway in melanoma. The hit compounds acquired in this investigation could have a significant impact on the development of novel medications that specifically target this cancer.
Materials and Methods
General
The present investigation utilized the following software: i) Autodock 4.2.6, ii) MGLTools 1.5.6, iii) Babel-2.4.0, iv) BIOVIA Discovery Studio 4.5, v) Avogadro Molecular Mechanics Force Fields (MMFF94), vi) Chemdraw Ultra 8.0, vii) Java Platform, and viii) SwissADME online tool. The SwissADME online tool was utilized to evaluate the pharmacokinetic characteristics of the ligands (http://www.swissadme.ch).
Ligand preparation
The ligands utilized in this work were obtained from the NPACT database (http://crdd.osdd.net/raghava/npact/) in SDF format. Subsequently, Babel-2.4.0 software was employed to convert the ligands to pdb format. The Avogadro software was then utilized to do energy minimization of the ligands. The ligands were modified by introducing hydrogen bonds and assigning rotatable bonds and Gasteiger charges using Autodock 4.2.6 software. The modified ligands were then saved in (Protein Data Bank, Partial Charge (Q), & Atom Type (T)) PDBQT format for subsequent docking procedures.
Protein preparation
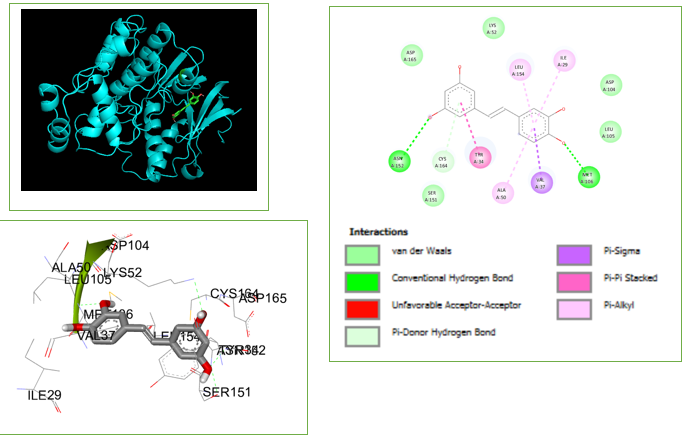
Structure of the ERK protein ([Figure 1]) was retrieved from protein data bank PDB (PDB ID: 2OJG).[4] The structure of 2OJG in complex with an inhibitor was prepared using Autodock 4.2.6 software, by adding polar hydrogens, merging non-polar hydrogens, assigning Kollmann charges, and saving the file in PDBQT format.
Molecular docking
Autodock Vina program was performed between the ligands and protein target for molecular docking analysis such as binding type, binding energy, inhibition activity, ligand efficiency, distance, and possible interactions. Their molecular docking scores were set as Autodock 4.2.6 tools and BIOVIA Discovery Studio software of the Molecular Graphics Laboratory software package by keeping the analogue flexible. [5], [6] The ligand efficiencies, binding energies, inhibition activity, hydrogen bonds, and bond lengths of the protein-ligand complexes were analysed using Autodock 4.2.6 software. BIOVIA Discovery Studio software was used for visual inspection of docking poses.[7]
Absorption, Distribution, Metabolism, Elimination & Toxicity (ADMET) Prediction
The pharmacokinetic and toxicity properties of the 9 compounds were achieved by using the SwissADME, which is an open online tool (http://www.swiss adme.ch). The ADME properties define blood-brain barrier (BBB) permeability and passive human gastrointestinal absorption (HIA) as well as a substrate or non-substrate permeability glycoprotein (P-gp) and Cytochrome P450 (CYP).
Results
|
S. No |
Ligand Name |
ΔG |
No of H bond |
H-Bond residues |
p-p stacked |
p-sigma |
p-alkyl |
|
1 |
Sulfurentin |
-9.2 |
1 |
ASP165 |
TYR34 |
LEU154 |
ILE29, VAL37 LYS52, GLN103, CYS164 |
|
2 |
Tectorigenin |
-8.8 |
3 |
ASP109, SER151 |
TYR34 |
- |
ILE29, VAL37, ALA50, LEU154 |
|
3 |
Parthenolide |
-8.7 |
2 |
SER151, ASN152 |
TYR34 |
- |
VAL37, ILE82, LEU154, CYS164 |
|
4 |
Epicatechin |
-8.6 |
2 |
ASP109, ASN152 |
- |
LEU154 |
ILE29, VAL37 |
|
5 |
Taiwaniaflavone |
-8.5 |
1 |
SER151 |
- |
- |
LEU154 |
|
6 |
Viscidulin II |
-8.6 |
3 |
SER151, ASN152, CYS164 |
- |
LEU154 |
VAL37, ALA50 |
|
7 |
Sumatrol |
-8.3 |
1 |
ASP165 |
TYR34 |
|
ILE29, ALA33, VAL37, CYS164 |
|
8 |
Piceatannol |
-8.2 |
2 |
MET106, ASN152 |
TYR34 |
VAL37 |
ILE29, LEU154 |
|
9 |
7-Methoxypraecansone B |
-7.5 |
3 |
LYS149, SER151, ASN152 |
TYR34 |
|
ILE29, VAL37, LEU154 |
|
10 |
Co-crystallized ligand (N,N-Dimethyl-4-(4-Phenyl-1h-Pyrazol-3-Yl)-1h-Pyrrole- 2- Carboxamide) [Redocked] |
-8.7 |
2 |
ASP104, MET106 |
- |
ILE29, TYR34, VAL37, LEU154 |
ALA50, CYS164 |
|
S.No |
Ligand Name |
RHB |
HBD |
HBA |
Mol.wt |
LogP |
|
1 |
Sulfurentin |
1 |
3 |
5 |
270.24 |
2.4196 |
|
2 |
Sumetrol |
3 |
1 |
7 |
410.422 |
3.4089 |
|
3 |
Taiwaniaflavone |
2 |
1 |
4 |
344.451 |
4.6422 |
|
4 |
Epicatechin |
1 |
5 |
6 |
290.271 |
1.5461 |
|
5 |
Parthenolide |
0 |
0 |
3 |
248.322 |
2.762 |
|
6 |
Piceatannol |
2 |
4 |
4 |
244.246 |
2.6794 |
|
7 |
Tectorigenin |
2 |
3 |
6 |
300.266 |
2.5854 |
|
8 |
Strychnine |
0 |
0 |
3 |
334.419 |
2.0925 |
|
9 |
Viscidulin II |
3 |
3 |
7 |
330.292 |
2.594 |
|
10 |
Methoxypraecansone B |
0 |
1 |
2 |
191.274 |
2.96802 |
Piceatannol, also known as trans-2,3′,4′, 5-tetrahydroxystilbene, is a phenolic molecule belonging to the stilbenoid class. It is a hydroxylated derivative of resveratrol. Piceatannol is derived from important sources such as grapes, passion fruit, white tea, Japanese knotweed, Asian legumes, and Korean rhubarb. Due to its lower concentration in grapes and wines compared to resveratrol, piceatannol has garnered significantly less scientific focus.[8]
Studying natural products, such as herbs and medicinal plants, is crucial due to their therapeutic potential and the important pharmacological compounds they contain.
These molecules have been optimized through the process of evolution to mimic pharmaceuticals and remain the main source of medications and potential therapeutic candidates. Out of a pool of 1574 compounds, we have chosen 464 natural compounds by applying Lipinski's rule of five. An assessment of the compound's druggability was conducted by utilizing online tools (Swiss ADME) to analyze its structure. An analysis was conducted on the features pertaining to the compound's suitability as a medication, and the findings are displayed in [Table 1]. The Autodock software was utilized to conduct molecular docking. The docking investigation revealed the presence of (PDB code: 2OJG). It's clear that both compounds have a strong interaction with MET 106 and ASN 152, as they form two hydrogen bond interactions ([Table 2]). [Figure 1] illustrate the maximum binding affinities that can occur between piceatannol and the 2OJG receptor binding sites. The molecular docking study showed that the chemical piceatannol had a stronger attraction to the ERK receptor's binding site. Based on the binding energy estimates, the new drug has a strong affinity for the binding pocket, which makes it very effective against the ERK receptor in melanoma.
Conclusion
Piceatannol has demonstrated favourable pharmacological characteristics, making it a promising candidate for the treatment of melanomas. This was identified using drug-likeness and ADMET predictions. The compound was determined to be non-toxic and had favourable interactions with the target protein. Consequently, its potential as an anticancer agent for the treatment or suppression of melanoma warrants further investigation in future experimental research.
Source of Funding
None
Conflicts of Interest
None
References
- Hu Y, Wu Y, Jiang C, Wang Z, Shen C, Zhu Z. Investigative on the Molecular Mechanism of Licorice Flavonoids Anti-Melanoma by Network Pharmacology, 3D/2D-QSAR, Molecular Docking, and Molecular Dynamics Simulation. Front Chem. 2022;10. [Google Scholar]
- Meier F, Schittek B, Busch S, Garbe C, Smalley K, Satyamoorthy K. The RAS/RAF/MEK/ERK and PI3K/AKT signaling pathways present molecular targets for the effective treatment of advanced melanoma. Front Biosci. 2005;10:2986-3001. [Google Scholar]
- Mangal M, Sagar P, Singh H, Raghava G, Agarwal S. NPACT: Naturally Occurring Plant-based Anti-cancer Compound-Activity-Target database. Nucleic Acids Res. 2013;41:1124-33. [Google Scholar]
- Aronov A, Baker C, Bemis G, Cao J, Chen G, Ford P. Flipped out: structure-guided design of selective pyrazolylpyrrole ERK inhibitors. J Med Chem. 2007;50(6):1280-7. [Google Scholar]
- Trott O, Olson A. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455-61. [Google Scholar]
- Yousuf M, Rafi S, Ishrat U, Shafiga A, Dashdamirova G, Leyla V. Potential Biological Targets Prediction, ADME Profiling, and Molecular Docking Studies of Novel Steroidal Products from Cunninghamella blakesleana. Med Chem. 2022;18(2):288-305. [Google Scholar]
- Riyaphan J, Pham D, Leong M, Weng C. Silico Approaches to Identify Polyphenol Compounds as α-Glucosidase and α-Amylase Inhibitors against Type-II Diabetes. . 2021;11(12). [Google Scholar]
- Banik K, Ranaware A, Harsha C, Nitesh T, Girisa S, Deshpande V. Piceatannol: A natural stilbene for the prevention and treatment of cancer. Pharmacol Res. 2020;153. [Google Scholar]
How to Cite This Article
Vancouver
Premkumar B, Yesuraj SR, Mohan S, Chandran S. Molecular docking studies for NPACT ligands for the treatment of melanoma skin cancer [Internet]. Int J Pharm Chem Anal. 2024 [cited 2025 Oct 15];11(1):51-54. Available from: https://doi.org/10.18231/j.ijpca.2024.007
APA
Premkumar, B., Yesuraj, S. R., Mohan, S., Chandran, S. (2024). Molecular docking studies for NPACT ligands for the treatment of melanoma skin cancer. Int J Pharm Chem Anal, 11(1), 51-54. https://doi.org/10.18231/j.ijpca.2024.007
MLA
Premkumar, B, Yesuraj, Samson Raj, Mohan, Santhosh, Chandran, Savitha. "Molecular docking studies for NPACT ligands for the treatment of melanoma skin cancer." Int J Pharm Chem Anal, vol. 11, no. 1, 2024, pp. 51-54. https://doi.org/10.18231/j.ijpca.2024.007
Chicago
Premkumar, B., Yesuraj, S. R., Mohan, S., Chandran, S.. "Molecular docking studies for NPACT ligands for the treatment of melanoma skin cancer." Int J Pharm Chem Anal 11, no. 1 (2024): 51-54. https://doi.org/10.18231/j.ijpca.2024.007
